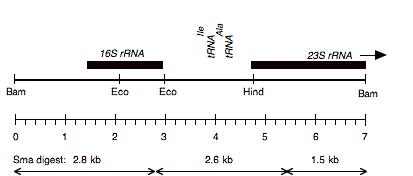Plasmids
Boynton-Gillham laboratory, Duke University, 1989
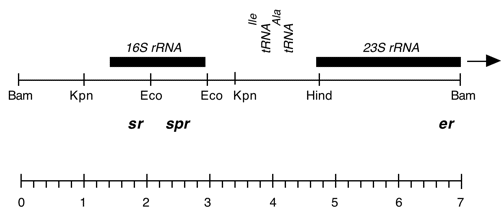
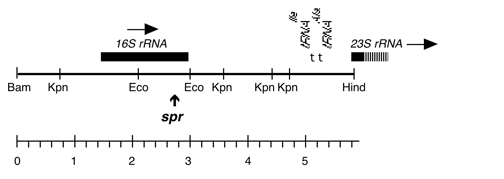
Chloroplast DNA BamHI 11/12 (7.0 kb) in pUC18; contains 16S and 5′ end of 23S rRNA genes, from CC-227, with sr-u-2-60, spr-u-1-6-2 and er-u-37 mutations
host strain: TG-1
amp resistant
More information
Strain: CC-227 sr-u-2-60 spr-u-1-6-2 er-u-37
Insert: BamHI 11/12 (7.0 kb)
Genes present: 16S rRNA, 23SrRNA (5’end)
Vector: pUC18
Bacterial host strain: TG-1
Selectable marker: amp-r, white on X-gal
Origin: Boynton-Gillham laboratory, 1988
see Harris et al., Genetics 123, 281 (1989) for mutations
P-202 cpDNA BamHI 6
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1989
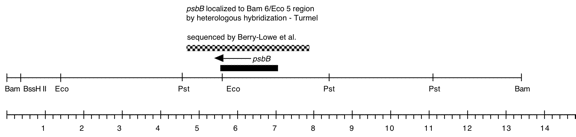
Chloroplast DNA BamHI 6 (13.5 kb) in pBR322; contains 3′ end of rps2 rpoA, psbB, psbN, and psbH; P-21 is the same insert in pBR313
host strain: DH1?
amp resistant
More information
Strain: CC-126 (a strain no longer in the stock collection)
CC-126 was derived from a cross of an ac20 cr1 double mutant to a wild-type strain.
It was a tetrad product with the wild-type alleles of both these mutations.
Insert: BamHI 6 (ca. 13.4 kb)
Genes present: rps2-C terminus (part), rpoA, pbB, psbT, psbN, psbH
Vector: pBR322
Bacterial host strain: DH1?
Selectable marker: amp-r
Origin: Jeff Woessner in Boynton/Gillham lab, 1990
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
As indicated above in the “Genes present” line, several other genes were subsequently identified in this fragment.
Boynton-Gillham laboratory, Duke University, 1989
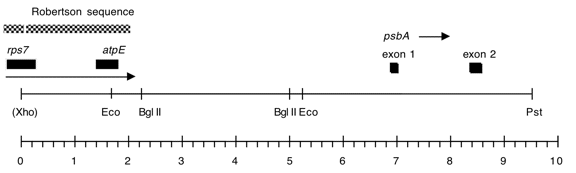
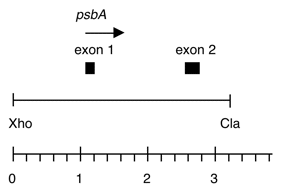
Chloroplast DNA PstI-XhoI (8.75 kb) in pUC18, from CC-125 wild type; contains atpE and 3′ end of rps7
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: PstI-(XhoI) SalI (ca. 9 kb); conflict in original data regarding size
This is the overlap between Pst 8 and Xho 5, cloned into the Pst-Sal sites of the plasmid.
The Xho site was lost in the cloning process.
Genes present: atpE, rps7 (3’end), psbA exons 1 and 2
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Niki Robertson in Boynton/Gillham lab, 1989
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
Also, the distance on the map is larger than the stated size of the insert.
Boynton-Gillham laboratory, Duke University, 1989
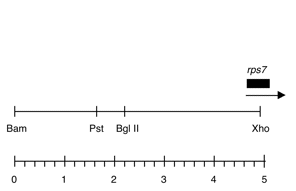
Chloroplast DNA BamHI-XhoI (5.0 kb) in pUC118, from CC-125 wild type; contains psbZ, psbM, rps14, and 5′ end of rps7
host strain: TG-1
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: BamHI-XhoI (5.0 kb)
Genes present: rps7 5′ end
Vector: pUC118
Bacterial host strain: TG-1
Selectable marker: amp-r, white on X-gal
Origin: Niki Robertson in Boynton-Gillham lab, 1989
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
Boynton-Gillham laboratory, Duke University, 1989
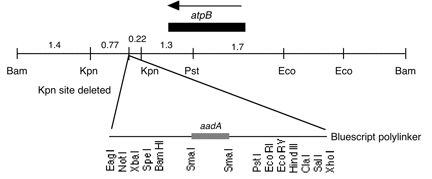
Chloroplast DNA BamHI-EcoRI (7.3 kb) in pUC8; contains aadA inserted downstream of atpB
host strain: JM83
amp resistant
More information
Strain: cw15
Insert: BamHI-EcoRI (7.3 kb)
Genes present: aadA, C. reinhardtii atpB
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton-Gillham lab, 1989
This plasmid was derived from one constructed in the laboratory of Lawrence Bogorad, in which the NPTII gene under control of the maize rbcL promoter was inserted downstream of atpB (Blowers et al. 1989, Plant Cell 1, 123-132). The NPTII-rbcL fragment was replaced with the E. coli aadA gene to create P-210.
Boynton-Gillham laboratory, Duke University, 1989
Chloroplast DNA EcoRI 17 (4.9 kb) in pUC8; contains rps12 with sr-u-sm2 mutation conferring streptomycin resistance, from CC-215
host strain: JM83
amp resistant
Liu XQ, Gillham NW, Boynton JE (1989) Chloroplast ribosomal protein gene rps12 of Chlamydomonas reinhardtii. Wild-type sequence, mutation to streptomycin resistance and dependence, and function in Escherichia coli. J Biol Chem 264:16100-16108
Boynton-Gillham laboratory, Duke University, 1989
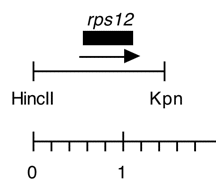
Chloroplast DNA HincII-KpnI (1.47 kb) in pUC119, from CC-125 wild type, subclone of EcoRI 17; contains wild-type rps12; P-175 is the same insert in pUC118
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: HincII-KpnI (1.47 kb)
Genes present: rps12
Vector: pUC119
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Paul Liu in Boynton/Gillham lab, 1989
Insert contains the 402 bp rps12 coding sequence plus 590 bp upstream and 480 bp downstream.
Reference: Liu et al., J. Biol. Chem. 264, 16100-16108 (1989).
P-214 contains the same insert cloned into pUC119
Liu XQ, Gillham NW, Boynton JE (1989) Chloroplast ribosomal protein gene rps12 of Chlamydomonas reinhardtii. Wild-type sequence, mutation to streptomycin resistance and dependence, and function in Escherichia coli. J Biol Chem 264:16100-16108
Boynton-Gillham laboratory, Duke University, 1989
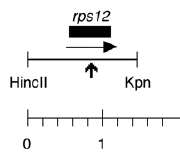
Chloroplast DNA HincII-KpnI (1.47 kb) in pUC18, from CC-125 wild type, subclone of EcoRI 17; contains rps12 in which Arg85 has been mutated to stop codon
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ (from plasmid P-70)
Insert: HincII-KpnI (1.47 kb)
Genes present: rps12
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Paul Liu in Boynton/Gillham laboratory, 1989
Insert = rps12 mutagenized in vitro (CGA arg at aa 85 changed to TGA stop codon) and cloned into SmaI site of pUC18. See Liu et al., J. Biol. Chem. 264, 16100 (1989).
Liu XQ, Gillham NW, Boynton JE (1989) Chloroplast ribosomal protein gene rps12 of Chlamydomonas reinhardtii. Wild-type sequence, mutation to streptomycin resistance and dependence, and function in Escherichia coli. J Biol Chem 264:16100-16108
Boynton-Gillham laboratory, Duke University, 1989
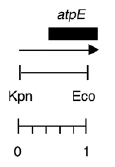
Chloroplast DNA EcoRI-KpnI (944 bp) in pUC119, from CC-125 wild type; contains 5′ end of atpE gene
host strain: TG-1
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: EcoRI-KpnI (944 bp)
Genes present: atpE 5′
Vector: pUC119
Bacterial host strain: TG-1
Selectable marker: amp-r, white on X-gal
Origin: Niki Robertson in Boynton/Gillham lab, 1989
This clone has the wild-type sequence of six A’s at positions 102-108 in the atpE transcript.
See Robertson et al., Mol. Gen. Genet. 221, 155-163 (1990)
Boynton-Gillham laboratory, Duke University, 1989
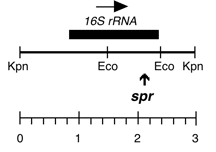
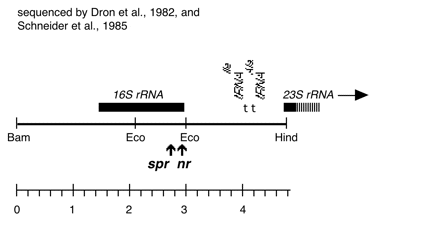
Chloroplast DNA Bam-HindIII (4.8 kb) in pUC18; 16S through 5′ part of 23S rRNA gene; from CC-110, contains spectinomycin resistance mutation spr-u-1-6-2; see P-667 for the same insert in an XL1 blue strain
host strain: JM83
amp resistant
More information
Strain: CC-110 spr-u-1-6-2 mt+
Insert: BamHI-HindIII (4.8 kb) from BamHI 11/12
Genes present: 16S rRNA, 23Ss rRNA (5′ end)
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1989
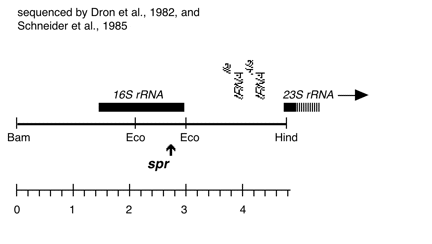
The spr-u-1-6-2 mutations is an A-> G change at bp 1123 in the C. reinhardtii 16S rRNA gene which causes loss of an AatII restriction site and confers high level spectinomycin resistance. See Harris et al., Genetics 123, 281-292 (1989). Used in rRNA transformation experiments by Newman et al., Genetics 126, 875-888 (1990).
Boynton-Gillham laboratory, Duke University, 1989
Chloroplast DNA BamHI-HindIII (5.9 kb) in pUC18, from C. smithii; contains 16S through 5′ end of 23S, with spectinomycin resistance mutation near 3′ end of 16S rRNA. The DNA in this strain came from CC-2477, which was a spectinomycin-resistant isolate from the C. smithii/C. reinhardtii hybrid strain CC-1852. The chloroplast genome in this strain was derived from C. smithii.
host strain: JM83
amp resistant
More information
Strain: CC-2477 C. smithii / C. reinhardtii hybrid
Insert: BamHI-HindIII (5.9 kb)
Genes present: 16S rRNA, 23S rRNA (5’end)
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1989
CC-2477 is a spectinomycin-resistant derivative of CC-1852, a hybrid between C. smithii (CC-1373) and C. reinhardtii (CC-124). The CC-124 parent was treated with acriflavine to eliminate mitochondrial DNA before mating, so CC-1852 and its derivatives contain the C. smithii versions of both chloroplast and mitochondrial DNA in a hybrid nuclear background. See Boynton et al. (1987) Proc. Natl. Acad. Sci. USA 84, 2391-2395 for derivation of the strain, and Newman et al. (1990) Genetics 126, 875-888 for use of CC-2477 in subsequent work.
The spectinomycin resistance mutation is an A-> G change at position 1123 in the C. reinhardtii 16S rRNA gene that confers high level spectinomycin resistance and destroys an AatII restriction site.
Newman SM, Boynton JE, Gillham NW, Randolph-Anderson BL, Johnson AM, Harris EH (1990) Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: molecular and genetic characterization of integration events. Genetics 126:875-888
Boynton-Gillham laboratory, Duke University, 1989
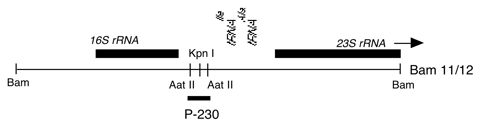
Chloroplast DNA 0.28 kb AatII fragment in pUC18, from BamHI 11/12, CC-125 wild type; spacer region between 16S and 23S rRNA genes; used as a probe for short dispersed repeat sequences in the chloroplast genome
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: AatII (0.28 kb)
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1990-
The insert is subclone of Bam 11/12, located within the spacer region between the 16S and 23S rRNA genes and containing short dispersed repeat elements. The AatII fragment was treated with Klenow to generate blunt ends and cloned into the SmaI site of pUC18. The AatII and SmaI sites have been lost but the insert can be excised using restriction sites in the pUC18 polylinker.
From Neil Hoffman to Boynton-Gillham laboratory, 1989
cDNA clone, 1.9 kb NcoI-BamHI fragment in pBSKS, containing nucleotides 619-2257 of the arylsulfatase gene
host strain: JM83
amp resistant
de Hostos EL, Schilling J, Grossman AR (1989) Structure and expression of the gene encoding the periplasmic arylsulfatase of Chlamydomonas reinhardtii. Mol Gen Genet 218:229-239
Boynton-Gillham laboratory, Duke University, 1990
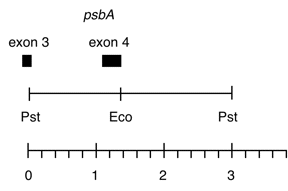
Chloroplast DNA PstI 17/18 (3.0 kb) in pUC8, from CC-125 wild type; contains psbA exon 4 and part of exon 3
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: PstI 17/18 (3.0 kb) [found to be slightly larger on analysis]
Genes present: psbA exons 3 and 4
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1990
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
Boynton-Gillham laboratory, Duke University, 1990
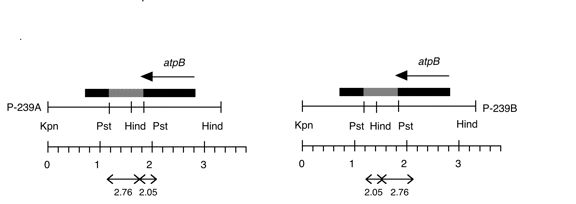
Chloroplast DNA KpnI-HindIII (3.3 kb) in pUC18, from a cw15 strain equivalent to CC-400; atpB disrupted with YEp24
host strain: JM83
amp resistant
More information
Strain: Rochaix cw15
Insert: KpnI-HindIII (3.3 kb)
Genes present: atpB (disrupted)
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: SELECTION: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1990
The insert contains a 0.48 kb PstI fragment from yeast plasmid YEp24 (P-238) cloned into the unique PstI site of the 2.8 kb KpnI-HindIII fragment (P-237), disrupting the atpB gene. P239A and B contain this Pst fragment inserted in opposite orientations. The plasmid was used in transformation experiments by Newman et al. (1991) Mol. Gen. Genet. 230, 65-74). See Figure 1 of that paper.
Newman SM, Gillham NW, Harris EH, Johnson AM, Boynton JE (1991) Targeted disruption of chloroplast genes in Chlamydomonas reinhardtii. Mol Gen Genet 230:65-74
Boynton-Gillham laboratory, Duke University, 1990
Chloroplast DNA KpnI (3.0 kb) in pUC18; 16S rRNA gene from C. smithii spectinomycin resistant mutant. The DNA in this strain came from CC-2477, which was a spectinomycin-resistant isolate from the C. smithii/C. reinhardtii hybrid strain CC-1852. The chloroplast genome in this strain was derived from C. smithii.
host strain: JM83
amp resistant
More information
Strain: CC-2477 C. smithii / C. reinhardtii hybrid
Insert: KpnI (3.0 kb)
Genes present: 16S rRNA
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1990
CC-2477 is a spectinomycin-resistant derivative of CC-1852, a hybrid between C. smithii (CC-1373) and C. reinhardtii (CC-124). The CC-124 parent was treated with acriflavine to eliminate mitochondrial DNA before mating, so CC-1852 and its derivatives contain the C. smithii versions of both chloroplast and mitochondrial DNA in a hybrid nuclear background. See Boynton et al. (1987) Proc. Natl. Acad. Sci. USA 84, 2391-2395 for derivation of the strain, and Newman et al. (1990) Genetics 126, 875-888 for use of CC-2477 in subsequent work.
The spectinomycin resistance mutation is an A-> G change at position 1123 in the C. reinhardtii 16S rRNA gene which confers high level spectinomycin resistance and destroys an AatII restriction site.
Boynton-Gillham laboratory, Duke University, 1990
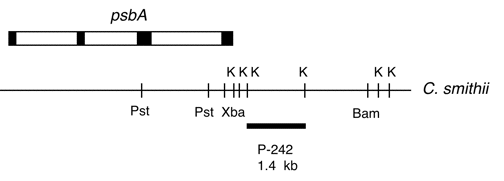
Chloroplast DNA KpnI (1.4 kb) in pUC18; from CC-1373 C. smithii, fragment from the spacer between psbA and 5S
host strain: JM83
amp resistant
More information
Strain: CC-1373 C. smithii wild type mt+
Insert: KpnI (1.4 kb)
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1990
The insert is from the spacer region between the psbA and 5S rRNA genes
Newman SM, Boynton JE, Gillham NW, Randolph-Anderson BL, Johnson AM, Harris EH (1990) Transformation of chloroplast ribosomal RNA genes in Chlamydomonas: molecular and genetic characterization of integration events. Genetics 126:875-888
Boynton-Gillham laboratory, Duke University, 1990
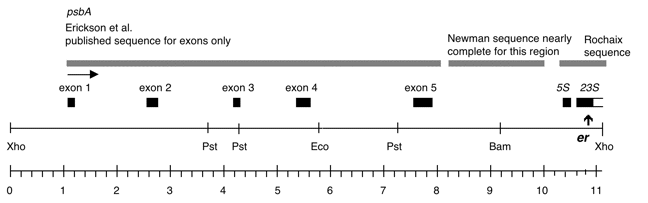
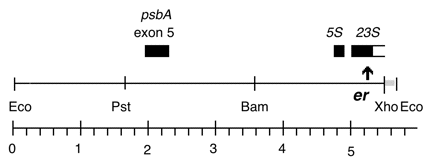
Chloroplast DNA XhoI 8 (11.0 kb) in BSKS+; contains psbA, 5S, and 3′ end of 23S with er-u-1a mutation from CC-64
host strain: JM109
amp resistant
More information
Strain: CC-64 er-u-1a mt+
Insert: XhoI 8 (11.0 kb)
Genes present: psbA, 5S rRNA, 23S rRNA (small exon)
Vector: BSKS+
Bacterial host strain: JM109
Selectable marker: amp-r, white on X-gal
Origin: Amnon Lers in Boynton/Gillham lab, 1990
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
The er-u-1a mutation is a C->T change at bp 2622 in C. reinhardtii 23S rRNA gene (Lemieux numbering, equivalent to E. coli 2611), which confers erythromycin resistance in Chlamydomonas. The insert was cloned in the XhoI site of the BSKS+ polylinker in both orientations. In P-259A, the psbA gene direction is from the KS SacI site towards the KS KpnI site; in P-259B, the psbA direction is from the KS KpnI site towards the KS SacI site.
Boynton-Gillham laboratory, Duke University, 1990
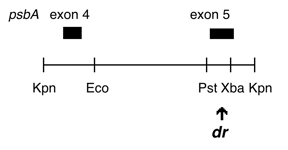
Chloroplast DNA KpnI (3.1 kb) in BSKS+; contains psbA exons 4 and 5 from DCMU4 mutant, CC-2473
host strain: JM109
amp resistant
More information
Strain: CC-2473 DCMU4
Insert: KpnI (3.1 kb)
Genes present: psbA exons 4 and 5
Vector: BSKS+
Bacterial host strain: JM109
Selectable marker: amp-r, white on X-gal
Origin: Amnon Lers in Boynton/Gillham lab, 1990
DCMU4 is a ser->ala mutation at aa 264 in the C. reinhardtii psbA 5th exon, conferring resistance to DCMU and atrazine. The insert was subcloned into the KS+ KpnI site in both orientations. In P-260A, the psbA direction is from the KS SacI site towards the KpnI site; in P-260B, it is from the KS KpnI site towards the SacI site.
Boynton-Gillham laboratory, Duke University, 1990
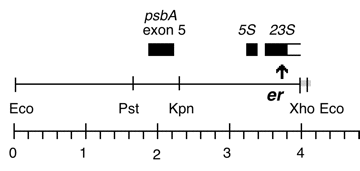
Chloroplast DNA EcoRI (5.51 kb) in pUC8 contains 3′ end of psbA, 5S, and 3′ exon of 23S with er-u-1a mutation from CC-64
host strain: JM109
amp resistant
More information
Strain: CC-64 er-u-1a mt+
Insert: EcoRI (5.51 kb)
Genes present: psbA 3′ end, 5S rRNA, 23S rRNA (small exon)
Vector: pUC8
Bacterial host strain: JM109
Selectable marker: amp-r, white on X-gal
Origin: Amnon Lers in Boynton/Gillham lab, 1990
The er-u-1a mutation is a C->T change at bp 2622 in the C. reinhardtii 23S rRNA gene (Lemieux numbering, equivalent to E. coli 2611), which confers erythromycin resistance in Chlamydomonas.
This is a subclone of P-259B. The insert contains sequences from the EcoRI site internal to psbA up to the EcoRI site in KS+, thus including some sequences of the SK+ polylinker beyond the original XhoI border of the insert in P-259B. Orientation of psbA is from the pUC8 Eco site toward the pUC8 HindIII site.
Boynton-Gillham laboratory, Duke University, 1990
Chloroplast DNA EcoRI (4.0 kb) in pUC8; contains 3′ end of psbA, 5S, and 3′ exon of 23S with er-u-1a mutation from CC-64; 1.5 kb deletion in spacer between psbA and 5S
host strain: JM109
amp resistant
More information
Strain: CC-64 er-u-1a mt+
Insert: EcoRI (4.0 kb)
Genes present: psbA (exon 5), 5S rRNA, 23S rRNA (small exon)
Vector: pUC8
Bacterial host strain: JM109
Selectable marker: amp-r, white on X-gal
Origin: Amnon Lers in Boynton/Gillham lab, 1990
This is a subclone of P-261. The three KpnI fragments including the BamHI site (approx. 1.5 kb) located between the psbA and 5S genes were deleted, and the remaining two Kpn ends were religated, leaving only one KpnI site. The insert appears slightly smaller than 4 kb.
The er-u-1a mutation is a C->T change at bp 2622 in C. reinhardtii 23S rRNA gene (Lemieux numbering, equivalent to E. coli 2611), which confers erythromycin resistance in Chlamydomonas.
Boynton-Gillham laboratory, Duke University, 1990
Chloroplast DNA BamHI-HindIII (4.8 kb) in pUC18; 16S rRNA gene from CC-245, containing the spr-u-1-H-4 and nr-u-2-1 mutations
host strain: JM83
amp resistant
More information
Strain: CC-245 nr-u-2-1 spr-u-1-H-4
Insert: BamHI-HindIII (4.8 kb)
Genes present: 16S rRNA, 23S rRNA (5’ end)
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1990
The nr-u-2-1 mutation is an A->G change at bp 1340 in the 16S rRNA gene, and the spr-u-1-H-4 mutations is an A->G change at bp 1123 in the same gene, conferring resistance to neamine/kanamycin and spectinomycin, respectively. See Harris et al., Genetics 123: 281-292 (1989).
Boynton-Gillham laboratory, Duke University, 1990
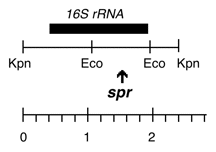
Chloroplast DNA KpnI (2.4 kb) in pUC18; 16S rRNA gene from CC-110, spectinomycin resistant mutant spr-u-1-6-2
host strain: JM83
amp resistant
More information
Strain: CC-110 spr-u-1-6-2 mt+
Insert: KpnI (2.4 kb)
Genes present: 16S rRNA
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1990
The spr-u-1-6-2 mutation is an A->G change at bp 1123 in the C. reinhardtii 16S rRNA gene which causes the loss of an AatII restriction site and confers high level spectinomycin resistance. See Harris et al., Genetics 123: 281-292 (1989).
Boynton-Gillham laboratory, Duke University, 1990
Chloroplast DNA ClaI (3.2 kb) in BSKS+; psbA 5′ flank, exons 1 and 2.
host strain: JM109
amp resistant
More information
Strain: CC-64 er-u-1a mt+
Insert: ClaI (3.2 kb)
Genes present: psbA 5’ flank, exons 1 and 2
Vector: BSKS+
Bacterial host strain: JM109
Selectable marker: amp-r, white on X-gal
Origin: Amnon Lers in Boynton/Gillham lab, 1990
The insert is a subclone of Xho8 cloned in BSKS+ (P-259) and contains sequences from ClaI to XhoI in the KS polylinker followed by psbA sequences from XhoI up to a ClaI site near exon 2 cloned into the ClaI site of BSKS+. Orientation of psbA is from KS Kpn site toward KS SacI site. Insert sequences in this fragment are identical to wild-typ
Boynton-Gillham laboratory, Duke University, 1990
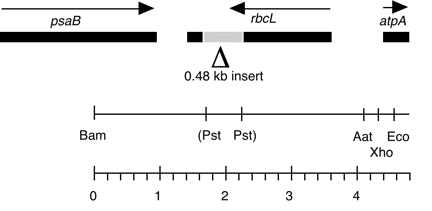
Chloroplast DNA BamHI-EcoRI (4.48 kb) in pUC18 with Pst site deleted, rbcL gene disrupted with YEp24. The DNA was obtained from CC-125 wild type.
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: BamHI-EcoRI (4.48 kb)
Genes present: rbcL, disrupted with 0.48 kb Pst I fragment from yeast plasmid YEp24
Vector: pUC18 with PstI site deleted
Bacterial host strain: JM83
Selectable marker: amp-r
Origin: Scott Newman in Boynton/Gillham lab, 1990
The insert contains the 0.48 kb Pst I fragment from yeast plasmid YEp24 (P-238), cloned into unique Pst site of a 4 kb Bam-Eco fragment of cpDNA, disrupting the rbcL gene. This construct was used in transformation experiments by Newman et al. (1991) Mol. Gen. Genet. 230, 65-74.
Newman SM, Gillham NW, Harris EH, Johnson AM, Boynton JE (1991) Targeted disruption of chloroplast genes in Chlamydomonas reinhardtii. Mol Gen Genet 230:65-74
Boynton-Gillham laboratory, Duke University, 1990
Chloroplast DNA BamHI 11/12 (7.0 kb) in pBR322, from CC-125 wild type; 16S through 5′ end of 23S; same insert as P-59
host strain: DH1?
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: BamHI 11/12 (7.0 kb)
Genes present: 16S rRNA, 23S rRNA (5’end)
Vector: pBR322
Bacterial host strain: DH1?
Selectable marker: amp-r
Origin: Jeff Woessner in Boynton/Gillham lab, 1990
See P-59 for the same insert in pUC8.
Boynton-Gillham laboratory, Duke University, 1990
Chloroplast DNA EcoRI 25 (3.1 kb) in pUC18, from CC-125 wild type; this fragment is internal to ORF2971
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: EcoRI 25 (3.1 kb)
Genes present: internal to ORF2971
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Barbara Randolph-Anderson in Boynton/Gillham lab, 1990
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-279A and P-279B have inserts cloned in opposite orientations.
Boynton-Gillham laboratory, Duke University, 1990
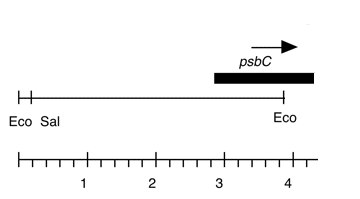
Chloroplast DNA EcoRI 20 (3.9 kb) in pUC18, from CC-125 wild type; contains the 3′ end of ORF2971 and the 5′ end of psbC
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: EcoRI 20 (3.9 kb)
Genes present: psbC (part)
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Barbara Randolph-Anderson in Boynton/Gillham lab, 1990
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
Boynton-Gillham laboratory, Duke University, 1990
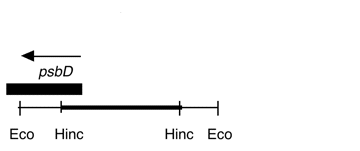
Chloroplast DNA HincII (1.5 kb) in pUC18, from a cw15 strain equivalent to CC-400; contains the 5′ end of psbD
host strain: JM83
amp resistant
More information
Strain: Rochaix cw15 strain
Insert: HincII (1.5 kb)
Genes present: psbD (5’ end)
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1990
This is a subclone of P-99, Eco 27.
Boynton-Gillham laboratory, Duke University, 1990
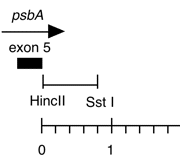
Chloroplast DNA HincII-SstI (0.8 kb) in pUC18, from CC-125 wild type; 3′ flank of the psbA gene
host strain: JM109
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: HincII-SstI (0.8 kb)
Vector: pUC18
Bacterial host strain: JM109
Selectable marker: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1990
- «Previous Page
- 1
- 2
- 3
- 4
- 5
- …
- 15
- Next Page»