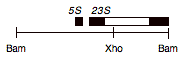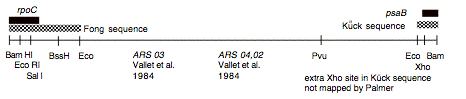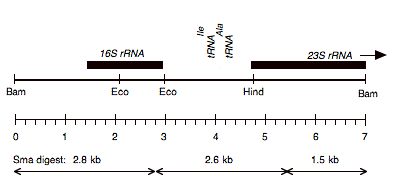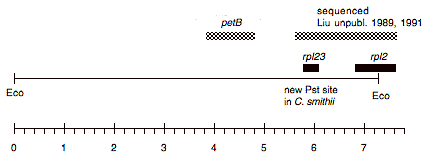Plasmids
MoClo Toolkit
$440.00
$440.00
The entire collection of plasmids, distributed as live cultures on two plates in a 96-well format, will be priced at $400 for academic/non-profit users and $800 for commercial/industry. Individual plasmids from the kit can also be ordered as bacterial cultures on agar plugs for standard plasmid pricing. You may view the individual list of plasmids in this collection here: https://www.chlamycollection.org/products/moclo-toolkit/.
From Crozet et al. (2018): “… we have developed a Modular Cloning toolkit for the green microalga Chlamydomonas reinhardtii. It is based on Golden Gate cloning with standard syntax, and comprises 119 openly distributed genetic parts, most of which have been functionally validated in several strains. It contains promoters, UTRs, terminators, tags, reporters, antibiotic resistance genes, and introns cloned in various positions to allow maximum modularity. The toolkit enables rapid building of engineered cells for both fundamental research and algal biotechnology. This work will make Chlamydomonas the next chassis for sustainable synthetic biology.”
Crozet et al. (2018) Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas reinhardtii. ACS Synthetic Biology 7:2074–2086
We are sorry but we cannot ship the MoClo Toolkit to China because of long shipping and customs delays. Individual clones can still be shipped however.
From Jean-David Rochaix to Boynton-Gillham laboratory, 1981
Chloroplast DNA BamH1 14/15 (3.0 kb) from Rochaix’s cw15 strain in pBR313; contains 3′ end of 23S rRNA gene through 5S; see P-124 for same insert in pBR322
host strain: C600
amp resistant
More information
Strain: Rochaix cw15
Insert: chloroplast DNA BamHI 14/15 (3.0 kb)
Genes present: 23S rRNA (3′ end), 5S rRNA
Vector: pBR313
Bacterial host strain: C600
Selectable marker: amp-r tet-s
Origin: Single cell clone of P-3 (Ba1 from Rochaix received 6/81)
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
From Jean-David Rochaix to Boynton-Gillham laboratory, 1981
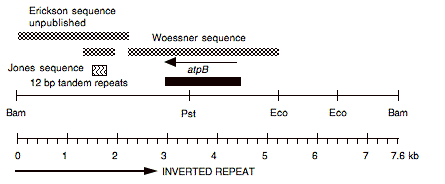
Chloroplast DNA BamHI 10 (7.6 kb) from Rochaix’s cw15 strain in pBR313; contains entire atpB gene and 3′ portion of ORF1995
host strain: C600
amp resistant
More information
Strain: Rochaix cw15
Insert: chloroplast DNA BamHI 10 (7.6 kb)
Genes present: atpB
Vector: pBR313
Bacterial host strain: C600
Selectable marker: amp-r tet-s
Origin: Single cell clone of P-6 (Ba 5 from Rochaix, received 6/81)
Reference: Boynton et al. (1988). See for use of this plasmid in transformation of atpB gene, and analysis of 12 bp tandem repeats./p>
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-19 cpDNA BamHI 8 (9.6 kb)
$30.00
$30.00
From Jean-David Rochaix to Boynton-Gillham laboratory, 1981
Chloroplast DNA BamHI 8 (9.6 kb) from Rochaix’s cw15 strain in pBR313; contains most of rpoC2 (3′) and 5′ end of psaB; see P-122 for same insert in pBR322
host strain: C600
amp resistant
Additional Information
Strain: Rochaix cw15
Insert: chloroplast DNA BamHI 8 (9.6 kb)
Genes present: rpoC2 (3′ end), psaB (extreme 5′ end), ARS04,02
Vector: pBR313
Bacterial host strain: C600
Selectable marker: amp-r tet-s
Origin: Single cell clone of P-8 (Ba7 from Rochaix, received 6/81)
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-21 cpDNA BamHI 6 (13.4 kb)
$30.00
$30.00
From Jean-David Rochaix to Boynton-Gillham laboratory, 1981
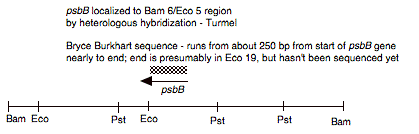
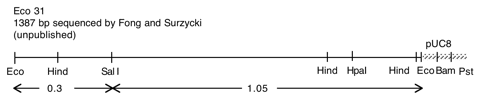
Chloroplast DNA BamHI 6 (13.4 kb) from Rochaix’s cw15 strain in pBR313; contains 3′ end of rps2 rpoA, psbB, psbN, and psbH; P-202 is the same insert in pBR322
host strain: C600
amp resistant
More information
Strain: Rochaix cw15
Insert: chloroplast DNA BamHI 6 (13.4 kb)
Genes present: psbH, psbN, ORF31, psbB, probably rps2
Vector: pBR313
Bacterial host strain: C600
Selectable marker: amp-r tet-s
Origin: Single cell clone of P-10 (Ba10 from Jean-David Rochaix, received 6/81
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
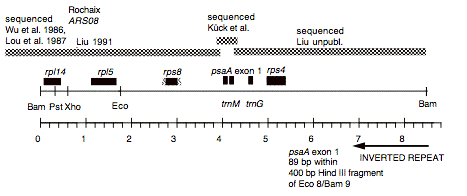
P-57 cpDNA BamHI 9 (8.4 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
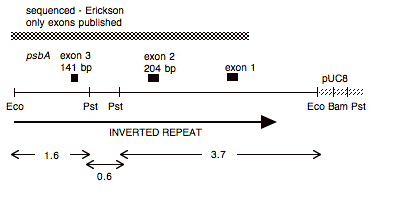
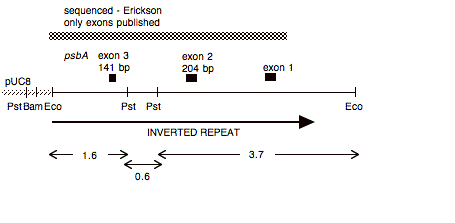
Chloroplast DNA BamHI 9 (8.4 kb) in pUC8, from CC-125 wild type; contains rpl14, rps8, psaA exon 1, and rps4
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA BamHI 9 (8.4 kb)
Genes present: rpl14, rpl5, rps8, psaA (exon 1), rps4
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA BamHI 11/12 (7.0 kb) in pUC8, from CC-125 wild type; contains 16S rRNA through 23S 5′ end
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA BamHI 11/12 (7.0 kb)
Genes present: 16S rRNA, 23s rRNA (5′ end)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
See P-271 for the same insert in pBR322.
P-62 cpDNA EcoRI 5 (10.5 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
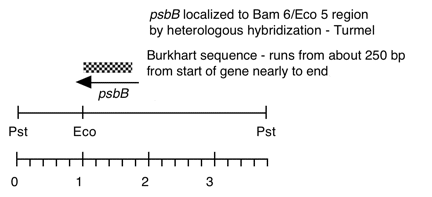
Chloroplast DNA EcoRI 5 (10.5 kb) in pUC8, from CC-125 wild type; contains psbB, rpoA, rps2 and most of rps18
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 5 (10.5 kb)
Genes present: psbB (5′), rps2, atpF
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Jeff Palmer in Boynton/Gillham lab, 1986
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
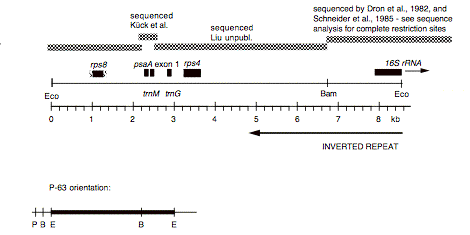
P-63 cpDNA EcoRI 8 (8.5 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 8 (8.5 kb) in pUC8, from CC-125 wild type; contains rps8, psaA exon 1, rps4 and 5′ end of 16S rRNA
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 8 (8.5 kb)
Genes present: rps8, psaA (exon 1), rps4, 16S rRNA (5′ end)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-64 cpDNA EcoRI 9 (7.6 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 9 (7.6 kb) in pUC8, from CC-125 wild type; contains clpP, rpl35, rpl23, and 5′ half of rpl2
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 9 (7.6 kb)
Genes present: clpP, trnL, petB, chlL, rpl23, rpl2 (5′)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-65 cpDNA EcoRI 12 (6.5 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 12 (6.5 kb) in pUC8, from CC-125 wild type; contains psaA exon 3
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 12 (6.5 kb)
Genes present: psaA (exon 3)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-66 cpDNA EcoRI 15 (6.0 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 15 (6.0 kb) in pUC8, from CC-125 wild type; contains psbA exons 1-4
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 15 (5.8 kb)psbA
Genes present: psbA exons 1-3
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
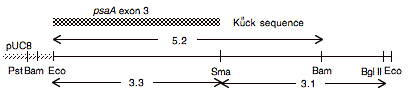
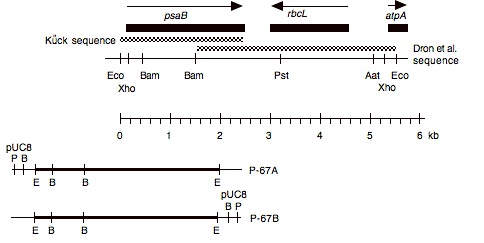
P-67 cpDNA EcoRI 14 (5.8 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 14 (5.8 kb) in pUC8, from CC-125 wild type; contains psaB, rbcL, and 5′ end of atpA
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 14 (5.5 kb)
Genes present: psaB, rbcL, atpA (5′)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-68 cpDNA EcoRI 15 (5.8 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 15 (5.8 kb) in pUC8, from CC-125 wild type; contains psbA exons 1-4
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 15 (5.8 kb) Gene psbA
Genes present: psbA (exons 1, 2, 3)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
Comment Same insert as P-66, but opposite orientation.
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
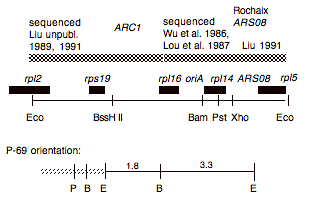
P-69 cpDNA EcoRI 16 (5.0 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 16 (5.0 kb) in pUC8, from CC-125 wild type; contains 3′ end of rpl2, rps19, rpl16, rpl14, and rpl5
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 16 (5.0 kb)
Genes present: rpl2 (part), rps19, rpl16, oriA, rpl14, rpl5
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
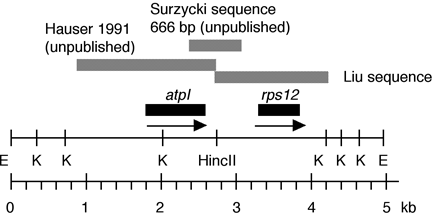
P-70 cpDNA EcoRI 17 (4.9 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 17 (4.9 kb) in pUC8, from CC-125 wild type; contains rps12, psaJ, atpI, and psbJ
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 17 (4.9 kb)
Genes present: psaJ, rps12
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-71 cpDNA EcoRI 18 (4.9 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 18 (4.9 kb) in pUC8, from CC-125 wild type; contains rps11, tscA, and most of chlN
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert:
Genes present:
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-72 cpDNA EcoRI 19 (4.4 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 19 (4.4 kb) in pUC8; contains psbH, psbN, and psbT
host strain: JM83
amp resistant
P-73 cpDNA EcoRI 24 (3.3 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 24 (3.3 kb) in pUC8, from CC-125 wild type; contains ycf12 and atpE
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 24 (3.3 kb)
Genes present: atpE
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab
Comment The 350 bp EcoRI-Bgl II subclone of this plasmid is a good probe for atpE.
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-74 cpDNA EcoRI 26 (3.0 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 26 (3.0 kb) in pUC8, from CC-125 wild type; internal to ORF2971
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 26 (3.0 kb)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-75 cpDNA EcoRI 31 (1.4 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 31 (1.4 kb) in pUC8, from CC-125 wild type; internal to rpoC2
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 31 (1.4 kb)
Genes present: rpoC2 (part)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-76 cpDNA EcoRI 32 (1.3 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA EcoRI 32 (1.3 kb) in pUC8, from CC-125 wild type; contains 3′ half of tufA
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 32 (1.3 kb)
Genes present: tufA (3′ end)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
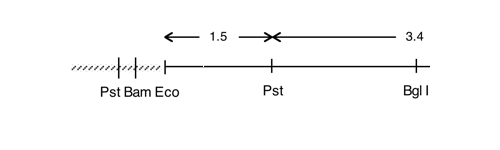
P-78 cpDNA PstI 4 (15.1 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA PstI 4 (15.1 kb) in pBR322, from CC-125 wild type; contains rps2, rps18, ycf3, ycf4, rps9, psbE, rpoB and psbF
host strain: SK2267
tetracycline resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA PstI 4 (15.1 kb)
Genes present: rps2 (probably), atpF, psbE, rpoB, psbF
Vector: pBR322
Bacterial host strain: SK2267
Selectable marker: tet-r, amp-s
Origin: David Lamb in Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
Orientation of P-78
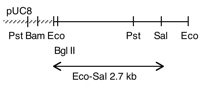
P-82 cpDNA PstI 14 (3.8 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA PstI 14 (3.8 kb) in pBR322, from CC-125 wild type; contains psbT and psbB
host strain: SK2267
tetracycline resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA PstI 14 (3.8 kb)
Genes present: psbB
Vector: pBR322
Bacterial host strain: SK2267
Selectable marker: tet-r, amp-s
Origin: David Lamb in Boynton/Gillham lab, 1983
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-84 cpDNA PstI 19 (2.9 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
Chloroplast DNA PstI 19 (2.9 kb) in pBR322, from CC-125 wild type; contains rpoC1b
host strain: SK2267
tetracycline resistant
Boynton-Gillham laboratory, Duke University, 1983
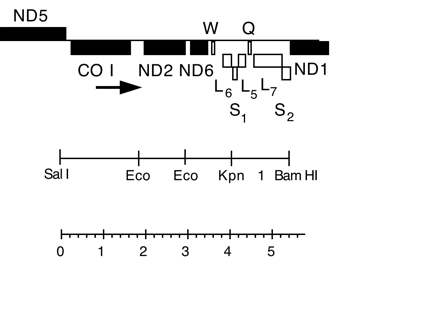
Mitochondrial Bam-Sal fragment (5.6 kb) in pBR322, from CC-125 wild type; contains cox1, nd2, nd6, W, L6, L7, Q, and S1
host strain: SK2267
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: mitochondrial DNA BamHI-SalI (5.6 kb)
Genes present: cox1, nd2, nd6, W, L6, L5, L7, Q, S1
Vector: pBR322
Bacterial host strain: SK2267
Selectable marker: amp-r
Origin: Boynton/Gillham lab, 1983
P-86 cpDNA EcoRI 22 (3.5 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
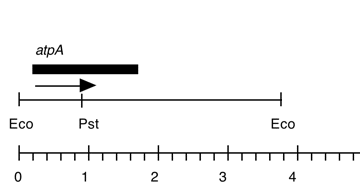
Chloroplast DNA EcoRI 22 (3.5 kb) in pUC8, from CC-125 wild type; contains nearly all of atpA, psbI, and most of cemA
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA EcoRI 22 (3.5 kb)
Genes present: atpA (most of gene; Eco site is near the 5′ end); psbI
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton-Gillham lab, 1983
Comment: This is a good probe for atpA.
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-87 cpDNA BamHI 7 (11.6 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1983
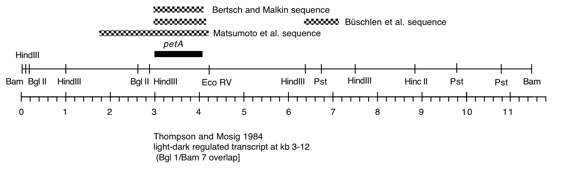
Chloroplast DNA BamHI 7 (11.6 kb) in pUC8, from CC-125 wild type; contains part of the Wendy transposon, petA, petD, and chlB
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+ 137c
Insert: chloroplast DNA BamHI 7 (11.6 kb)
Genes present: petA, petD, chlB
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r
Origin: Boynton/Gillham lab, 1985
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
Boynton-Gillham laboratory, Duke University, 1983
Nuclear rDNA unit, 3.6 kb BamH1 fragment in pUC8, from CC-125 wild type; contains part of 25S, 5.8S, and most of 18S rDNA
For map, see Marco and Rochaix (1980) Mol. Gen. Genet. 177, 715-723
host strain: JM83
amp resistant
Marco Y, Rochaix JD (1980) Organization of the nuclear ribosomal DNA of Chlamydomonas reinhardii. Mol Gen Genet 177:715-23
P-99 cpDNA EcoRI 27 (2.7 kb)
$30.00
$30.00
From Jean-David Rochaix to Boynton-Gillham laboratory, 1985
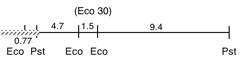
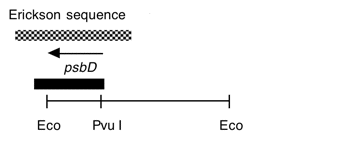
Chloroplast DNA EcoRI 27 (2.7 kb) from Rochaix’s cw15 strain in pCR1; contains most of psbD and the 5′ end of ORF2971
host strain: JM83
kanamycin resistant
More information
Strain: Rochaix cw15
Insert: chloroplast DNA EcoRI 27 (2.7 kb)
Genes present: psbD
Vector: pCR1 (11.4 kb)
Bacterial host strain: Uncertain
Selectable marker: kan-r
Origin: Jean-David Rochaix, 1985, as R3
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
- 1
- 2
- 3
- …
- 15
- Next Page»