Plasmids
Boynton-Gillham laboratory, Duke University, 1986
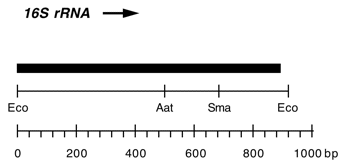
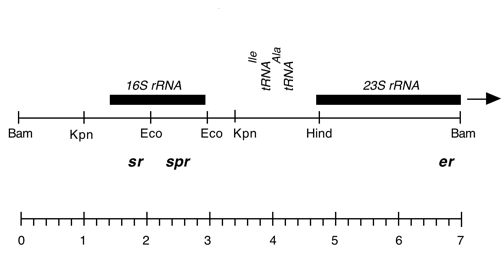
Chloroplast DNA EcoRI 36/37 (0.9 kb) in pUC8, from CC-125 wild type; contains wild-type 16S rRNA gene (3′ end);
host strain: JM83
amp resistant
More information
Strain: CC-125
Insert: EcoRI 36/37 (0.9 kb)
Genes present: 16S rRNA (3′ end)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1986
Harris EH, Burkhart BD, Gillham NW, Boynton JE (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292
Boynton-Gillham laboratory, Duke University, 1986
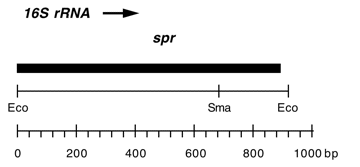
Chloroplast DNA EcoRI 36/37 (0.9 kb) in pUC8; contains 16S rRNA gene (3′ end) from spectinomycin resistant mutant CC-110 spr-u-1-6-2
host strain: JM83
amp resistant
More information
Strain: CC-110 spr-u-1-6-2 mt+
Insert: EcoRI 36/37 (0.9 kb)
Genes present: 16S rRNA (3′ end)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1986
Contains A->G change at bp 1123 in C. reinhardtii 16S rRNA gene, causing loss of an Aat II site and conferring high level spectinomycin resistance.
Harris et al., Genetics 123, 281 (1989).
Harris EH, Burkhart BD, Gillham NW, Boynton JE (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292
Boynton-Gillham laboratory, Duke University, 1986
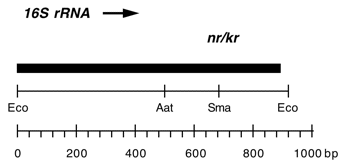
Chloroplast DNA EcoRI 36/37 (0.9 kb) in pUC8; contains 16S rRNA gene (3′ end) from CC-187, kanamycin resistant mutant kr-u-24-2
host strain: JM83
amp resistant
More information
Strain: CC-187 kr-u-24-2 mt+
Insert: EcoRI 36/37 (0.9 kb)
Genes present: 16S rRNA (3′ end)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1986
Contains C->T change at bp 1341 in C. reinhardtii 16S rRNA gene, conferring neamine and kanamycin resistance.
Harris et al., Genetics 123, 281 (1989).
Harris EH, Burkhart BD, Gillham NW, Boynton JE (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292
Boynton-Gillham laboratory, Duke University, 1986
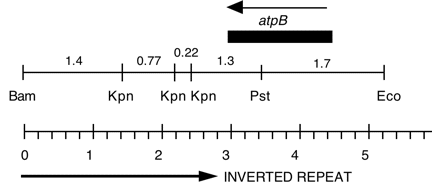
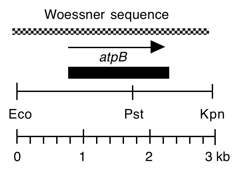
Chloroplast DNA BamHI-EcoRI (5.3 kb) in pUC8, from a cw15 strain equivalent to CC-400. This fragment is the overlap of Bam 10 and Eco 11 and contains the entire atpB gene.
host strain: JM83
amp resistant
More information
Strain: cw15, equivalent to CC-400
Insert: BamHI-EcoRI (5.3 kb), overlap of Bam 10 and Eco 11
Genes present: atpB
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton-Gillham lab, 1990
Woessner et al., Gene 44, 17-28 (1986)
P-119 cpDNA RsaI (700 bp)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1986
Chloroplast DNA 700 bp RsaI fragment in pUC8 from Kpn-Eco region of Bam 10, from a cw15 strain equivalent to CC-400; close to atpB but apparently not within the gene
host strain: JM83
amp resistant
P-122 cpDNA BamHI 8 (9.6 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1986
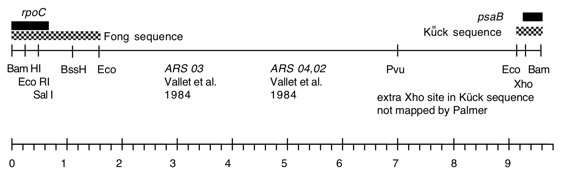
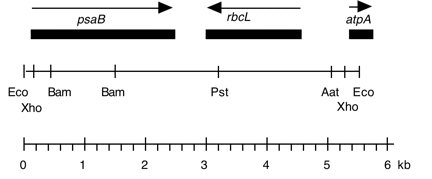
Chloroplast DNA BamHI 8 (9.6 kb) in pBR322, from CC-125 wild type; contains most of rpoC2 (3′) and 5′ end of psaB; see P-19 for same insert in pBR313
host strain: DH1
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: BamHI 8 (9.6 kb)
Genes present: rpoC (part) , psaB (part)
Vector: pBR322
Bacterial host strain: DH1
Selectable marker: amp-r
Origin: Jeff Woessner in Boynton/Gillham lab, 1990
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
Boynton-Gillham laboratory, Duke University, 1986
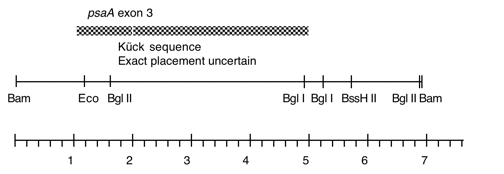
Chloroplast DNA BamHI 13 (6.8 kb) in pBR322, from CC-125 wild type; contains most of ccsA and psaA exon 3
host strain: DH1
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: BamHI 13 (6.8 kb)
Genes present: psaA exon 3
Vector: pBR322
Bacterial host strain: DH1
Selectable marker: amp-r
Origin: Jeff Woessner in Boynton/Gillham lab, 1990
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
Boynton-Gillham laboratory, Duke University, 1986
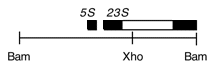
Chloroplast DNA BamHI 14/15 (3.0 kb) in pBR322, from CC-125 wild type; contains 3′ end of 23S rRNA gene through 5S; see P-14 for same insert in pBR313
host strain: DH1
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: BamHI 14/15 (3.0 kb)
Genes present: 23S rRNA (3′ end), 5S rRNA
Vector: pBR322
Bacterial host strain: DH1
Selectable marker: amp-r
Origin: Jeff Woessner in Boynton/Gillham lab, 1990
Boynton-Gillham laboratory, Duke University, 1986
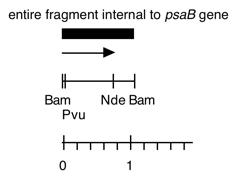
Chloroplast DNA BamHI 16 (1.1 kb) in pBR322, from CC-125 wild type; internal to psaB
host strain: DH1
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: BamHI 16 (1.1 kb)
Genes present: psaB
Vector: pBR322
Bacterial host strain: DH1
Selectable marker: amp-r
Origin: Jeff Woessner in Boynton/Gillham lab, 1990
This map is from the original file for the plasmid and was drawn before the complete sequence was known.
Boynton-Gillham laboratory, Duke University, 1986
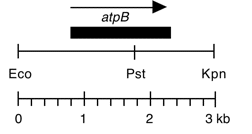
Chloroplast DNA EcoRI-KpnI (3.0 kb) in pUC18, from CC-125 wild type; contains atpB
host strain: JM103
amp resistant
More informatin
Strain: CC-125 wild type mt+
Insert: KpnI-EcoRI (3.0 kb)
Genes present: atpB
Vector: pUC18
Bacterial host strain: JM103
Selectable marker: amp-r, white on X-gal
Origin: Jeff Woessner in Boynton-Gillham lab, 1990
Boynton-Gillham laboratory, Duke University, 1986
Chloroplast DNA EcoRI-KpnI (3.0 kb) in pUC18; atpB from CC-440, containing the ac-u-c-2-29 mutation (K154N)
host strain: JM103
amp resistant
More information
Strain: C. reinhardtii CC-440 ac-u-c-2-29
Insert: KpnI-EcoRI (3.0 kb)
Genes present: atpB
Vector: pUC18
Bacterial host strain: JM103
Selectable marker: amp-r, white on X-gal
Origin: Jeff Woessner in Boynton/Gillham lab, 1990
Robertson D, Woessner JP, Gillham NW, Boynton JE (1989) Molecular characterization of two point mutants in the chloroplast atpB gene of the green alga Chlamydomonas reinhardtii defective in assembly of the ATP synthase complex. J Biol Chem 264:2331-2337
Boynton-Gillham laboratory, Duke University, 1986
Chloroplast DNA 900 bp HindIII fragment in pUC9, from CC-125 wild type; internal to rbcL
host strain: JM83
amp resistant
P-149 RBCS2 EcoRI (0.8 kb)
$30.00
$30.00
From Jean-David Rochaix to Boynton-Gillham laboratory, 1987
Nuclear DNA, 0.8 kb EcoRI fragment from Rochaix’s cw15 strain in pUC18; CS2.1 probe for RBCS2
host strain: JM83
amp resistant
Goldschmidt-Clermont M, Rahire M (1986) Sequence, evolution and differential expression of the two genes encoding variant small subunits of ribulose bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. J Mol Biol 191:421-432
From Don Weeks to Boynton-Gillham laboratory, 1985
3.1 kb XhoI-SmaI fragment in pUC9, probe for alpha-1 tubulin
host strain: JM83
amp resistant
From Don Weeks to Boynton-Gillham laboratory, 1985
3.1 kb XhoI fragment in pBR322, beta-2 tubulin + 3′ flank
host strain: JM83
amp resistant
P-156 cpDNA BamHI 4 (20 kb)
$30.00
$30.00
From Jean-David Rochaix to Boynton-Gillham laboratory, 1987
Chloroplast DNA BamHI 4 (20 kb) in pUC18, from Rochaix’s cw15 strain; contains psbZ, psbM, rps14, rps7, atpE, ycf12, and psbA; see P-50 for same insert in pUC8
host strain: 520
amp resistant
From Jean-David Rochaix to Boynton-Gillham laboratory, 1987
Chloroplast DNA BamHI-BglII 10 kb fragment in pUC18; subclone of Bam 4; psbA from DCMU4 mutant (= CC-2473).
host strain: 521
amp resistant
More information
Strain: DCMU4, equivalent to CC-2473
Insert: BamHI-BglI (10 kb)
Note: Bam-Bgl digests consistently yield a small amount of vector + insert, with most of the DNA uncut
Genes present: psbA
Vector: pUC18
Bacterial host strain: 521
Selectable marker: amp-r, white on X-gal
Origin: Jean-David Rochaix to Boynton/Gillham lab, 1987
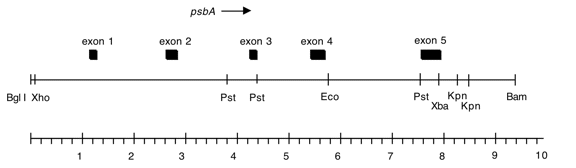 See Erickson et al., PNAS 81, 3617 (1984)
See Erickson et al., PNAS 81, 3617 (1984)
C. reinhardtii DCMU4 contains a ser->ala alteration in the 5th exon of psbA at position 264 conferring resistance to DCMU and atrazine.
From René Matagne to Boynton-Gillham laboratory, 1987
Mitochondrial BamHI-SalI fragment (0.3 kb) in pBR322; from C. smithii (equivalent to CC-1373); = Matagne pULG-S2
host strain: HB101
amp resistant
Boynton-Gillham laboratory, Duke University, 1987
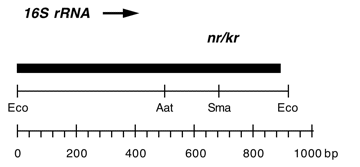
Chloroplast DNA EcoRI 36/37 (0.9 kb) in pUC8; contains 16S rRNA gene (3′ end) from CC-86 neamine/kanamycin resistant mutant nr-u-2-1
host strain: JM83
amp resistant
More information
Strain: CC-86 nr-u-2-1 mt+
Insert: EcoRI 36/37 (0.9 kb)
Genes present: 16S rRNA (3′ end)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1987
Contains A->G at bp 1340 in C. reinhardtii 16S rRNA gene which produces high level resistance to neamine and kanamycin.
Harris et al., Genetics 123, 281 (1989).
Harris EH, Burkhart BD, Gillham NW, Boynton JE (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292
Boynton-Gillham laboratory, Duke University, 1987
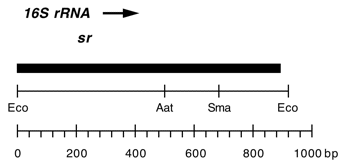
Chloroplast DNA EcoRI 36/37 (0.9 kb) in pUC8; contains 16S rRNA gene (3′ end) from CC-267 streptomycin resistant mutant sr-u-sm5
host strain: JM83
amp resistant
More information
Strain: CC-267 sr-u-sm5
Insert: EcoRI 36/37 (0.9 kb)
Genes present: 16S rRNA (3′ end)
Vector: pUC8
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1987
Contains A->C change at bp 858 in C. reinhardtii 16S rRNA gene which produces high level of streptomycin resistance.
Harris et al., Genetics 123, 281 (1989).
Harris EH, Burkhart BD, Gillham NW, Boynton JE (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292
P-169 ATPC EcoRI (1.9 kb)
$30.00
$30.00
From Bruce Selman and Lloyd Yu to Boynton-Gillham laboratory, 1987
1.9 kb EcoRI fragment in pTZ18R; ATPC, encoding the gamma subunit of CF1 chloroplast ATP synthase
host strain: JM101
amp resistant
Yu LM, Merchant S, Theg SM, Selman BR (1988) Isolation of a cDNA clone for the gamma subunit of the chloroplast ATP synthase of Chlamydomonas reinhardtii: import and cleavage of the precursor protein.Proc Natl Acad Sci USA 85:1369-1373
From Robert Spreitzer to Boynton-Gillham laboratory, 1988
Chloroplast DNA EcoRI 14 (5.8 kb) in pBR329; rbcL containing Spreitzer 68-4PP ts mutation (L290F).
host strain: JM83
amp resistant
More information
Strain: CC-2338 68-4PP, rbcL mutant
Insert: EcoRI 14 (5.8 kb)
Genes present: psaB, rbcL, atpA (part)
Vector: pBR329
Bacterial host strain: JM83
Selectable marker: amp-r, tet-r
Origin: Bob Spreitzer, 1988 to Boynton/Gillham lab
The 68-4PP mutant has a ts photosynthesis deficient phenotype due to a phe for leu substitution at residue 290 in the Rubisco large subunit.
References:
Chen et al. (1993) Plant Physiol 101, 1189-1194
Chen et al. (1988) Proc. Natl. Acad. Sci. USA 85, 4696-4699
Boynton-Gillham laboratory, Duke University, 1988
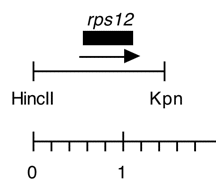
Chloroplast DNA 1.47 kb HincII-KpnI fragment in pUC118, from CC-125 wild type; subclone of Eco 17; contains rps12
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: HincII-KpnI (1.47 kb)
Genes present: rps12
Vector: pUC118
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1988
Insert contains the 402 bp rps12 coding sequence plus 590 bp upstream and 480 bp downstream.
Reference: Liu et al., J. Biol. Chem. 264, 16100-16108 (1989).
Liu XQ, Gillham NW, Boynton JE (1989) Chloroplast ribosomal protein gene rps12 of Chlamydomonas reinhardtii. Wild-type sequence, mutation to streptomycin resistance and dependence, and function in Escherichia coli. J Biol Chem 264:16100-16108
Boynton-Gillham laboratory, Duke University, 1988
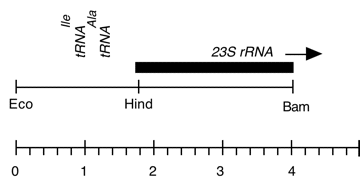
Chloroplast DNA BamHI-EcoRI (4.1 kb) in pUC18, from CC-125 wild type; 5′ end of 23S rRNA gene (wild type), overlap of Eco 3/4 and Bam 11/12
host strain: TG-1
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: BamHI-EcoRI (4.1 kb)
overlap of Eco 3/4 and Bam 11/12
Genes present: 23S rRNA (5′ end)
Vector: pUC18
Bacterial host strain: TG-1
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1988
Boynton-Gillham laboratory, Duke University, 1988
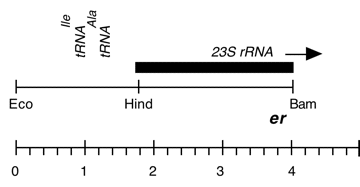
Chloroplast DNA BamHI-EcoRI (4.1 kb) in pUC18; 5′ end of 23S rRNA gene from CC-67 erythromycin resistant mutant er-u-37
host strain: TG-1
amp resistant
More information
Strain: CC-67 er-u-37
Insert: BamHI-EcoRI (4.1 kb)
overlap of Eco 3/4 and Bam 11/12
Genes present: 23S rRNA (5′ end)
Vector: pUC18
Bacterial host strain: TG-1
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1988
The er-u-37 mutation is a G->A change at bp 2066 in C. reinhardtii 23S rRNA
(corresponding to E. coli 2057) which produces resistance to erythromycin.
See Harris et al., Genetics 123, 281 (1989).
Harris EH, Burkhart BD, Gillham NW, Boynton JE (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292
Boynton-Gillham laboratory, Duke University, 1988
Chloroplast DNA BamHI 11/12 (7.0 kb) in pUC18; 16S through 5′ end of 23S rRNA gene; contains sr-u-2-60, spr-u-1-6-2, and er-u-37 mutations
host strain: JM83
amp resistant
More information
Strain: CC-227 spr-u-1-6-2 sr-u-2-60 er-u-37
Insert: BamHI 11/12 (7.0 kb)
Genes present: 16S rRNA, 23S rRNA (5’end)
Vector: pUC18
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1988
See Harris et al., Genetics 123, 281 (1989) for mutations, Newman et al., Genetics 126, 875 (1990) for use of this plasmid in transformation experiments.
Note: by selecting cells transformed with this plasmid simultaneously for spectinomycin and streptomycin or erythromycin resistance one essentially eliminates the possibility of recovering spontaneous antibiotic resistant mutants.
However, cells carrying both spr-u-1-6-2 and sr-u-2-60 have been found to be deficient in chloroplast protein synthesis and have impaired photosynthetic performance under high iradiance conditions, consistent with reduced levels of D1 synthesis.
Thus if selection for antibiotic resistance is to be used specifically for cotransformation, we recommend using P-228 (spr-u-1-6-2 alone) instead.
Steve Mayfield to Boynton-Gillham laboratory, 1989
EcoRI-KpnI (8.0 kb) in pUC19; genomic fragment from FUD44 mutant (equivalent to CC-2892) in the PSBO gene encoding the OEE1 protein
host strain: JM83
amp resistant
Mayfield SP, Kindle KL (1990) Stable nuclear transformation of Chlamydomonas reinhardtii by using a C. reinhardtii gene as the selectable marker. Proc Natl Acad Sci USA 87:2087-2091
Boynton-Gillham laboratory, Duke University, 1989
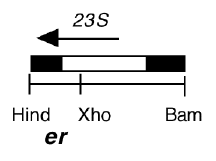
Chloroplast DNA BamHI-HindIII (1.6 kb) in pUC119; 3′ end of 23S rRNA gene from CC-64, erythromycin resistant mutant er-u-1a
host strain: probably TG-1
amp resistant
More information
Strain: CC-64 er-u-1a
Insert: BamHI-HindIII (1.6 kb)
Genes present: 23S rRNA (3’end)
Vector: pUC119
Bacterial host strain: probably TG-1
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1988
The mutation in er-u-1a is a C->T change at bp 2622 in the C. reinhardtii 23S rRNA gene (corresponding to E. coli 2611), which produces erythromycin resistance.
See Harris et al., Genetics 123, 281 (1989).
Harris EH, Burkhart BD, Gillham NW, Boynton JE (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292
Boynton-Gillham laboratory, Duke University, 1989
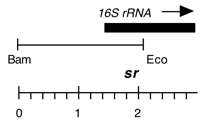
Chloroplast DNA EcoRI-BamHI (2.0 kb) in pUC8; 5′ end of 16S rRNA gene; contains streptomycin resistance mutation sr-u-2-60, from CC-118
host strain: TG-1
amp resistant
More information
Strain: CC-118 sr-u-2-60
Insert: EcoRI-BamHI (2.0 kb), subclone of Bam 11/12
Genes present: 16S rRNA (5′ end)
Vector: pUC8
Bacterial host strain: TG-1
Selectable marker: amp-r, white on X-gal
Origin: Boynton/Gillham lab, 1990
The sr-u-2-60 mutation is an A->C change at base 474 in C. reinhardtii 16S rRNA gene that produces streptomycin resistance.
See Harris et al., Genetics 123, 281 (1989).
Harris EH, Burkhart BD, Gillham NW, Boynton JE (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292
Boynton-Gillham laboratory, Duke University, 1989
Chloroplast DNA EcoRI-BamHI (2.0 kb) in pUC8; 5′ end of 16S rRNA gene; contains streptomycin resistance mutation sr-u-sm3, from CC-266
host strain: TG-1
amp resistant
Harris EH, Burkhart BD, Gillham NW, Boynton JE (1989) Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics 123:281-292
- «Previous Page
- 1
- 2
- 3
- 4
- …
- 15
- Next Page»