Plasmids
P-505 cpDNA EcoRI 34
$30.00
$30.00
From Saul Purton, 1993
Chloroplast DNA EcoRI 34 (ca. 1.1 kb) in pUC9; contains psaA exon 2 and 3′ end of psbD
host strain: unknown
amp resistant
P-520 cpDNA PstI 11 (7.8 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke Univesrsity, 1993
Chloroplast DNA PstI 11 (7.8 kb) in BSKS+; contains atpA (3′ end), atpH, rps11, rpoA
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: PstI 11 (7.8 kb)
Genes present: atpA (part), atpH, rpoA (part)
Vector: BSKS+
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Barbara Randolph-Anderson in Boynton/Gillham lab, 1993
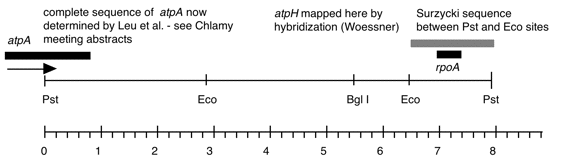
This plasmid was derived from P-80, which was the same insert in a tetracycline-resistant vector. This map is from the original file for plasmid P-80 and was drawn before the complete sequence was known.
P-521 cpDNA PstI 13 (4.0 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke Univesrsity, 1993
Chloroplast DNA PstI 13 (4.0 kb) in BSKS+; internal to ORF2971
host strain: JM83
amp resistant
More information
Strain: CC-125 wild type mt+
Insert: PstI 13 (4.0 kb)
Genes present: internal to ORF2971
Vector: BSKS+
Bacterial host strain: JM83
Selectable marker: amp-r, white on X-gal
Origin: Barbara Randolph-Anderson in Boynton/Gillham lab, 1993

This plasmid was derived from P-81, which was the same insert in a tetracycline-resistant vector. This map is from the original file for the plasmid and was drawn before the complete sequence was known.
P-537 ATPC cDNA
$30.00
$30.00
From Sabeeha Merchant, 1993
ATPC cDNA (CF1 ATP synthase, gamma subunit), EcoRI (1.8 kb) in pTZ18R
host strain: JM101
amp resistant
Yu LM, Merchant S, Theg SM, Selman BR (1988) Isolation of a cDNA clone for the gamma subunit of the chloroplast ATP synthase of Chlamydomonas reinhardtii: import and cleavage of the precursor protein.Proc Natl Acad Sci USA 85:1369-1373
P-538 cytochrome c6 cDNA
$30.00
$30.00
From Sabeeha Merchant, 1993
Cytochrome c6 cDNA, 710 bp EcoRI fragment in pGEM1
host strain: HB101
amp resistant
Merchant S, Bogorad L (1987) The Cu(II)-repressible plastidic cytochrome c. Cloning and sequence of a complementary DNA for the pre-apoprotein. J Biol Chem 262:9062-9067
P-539 plastocyanin cDNA
$30.00
$30.00
From Sabeeha Merchant, 1993
Plastocyanin cDNA, 577 bp EcoRI fragment in pTZ18R
host strain: HB101
amp resistant
Merchant S, Hill K, Kim JH, Thompson J, Zaitlin D, Bogorad L (1990) Isolation and characterization of a complementary DNA clone for an algal pre-apoplastocyanin. J Biol Chem 265:12372-12379
P-540 cytochrome c6 genomic
$30.00
$30.00
From Sabeeha Merchant, 1993
Cytochrome c6 genomic, ca. 5 kb SstI fragment in pTZ19R
host strain: JM101
amp resistant
Hill KL, Li HH, Singer J, Merchant S (1991) Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J Biol Chem 266:15060-15067
P-541 plastocyanin genomic
$30.00
$30.00
From Sabeeha Merchant, 1993
Plastocyanin genomic, ca. 6.5 kb KpnI fragment in pTZ19R
host strain: DH5 alpha
amp resistant
Quinn J, Li HH, Singer J, Morimoto B, Mets L, Kindle K, Merchant S (1993) The plastocyanin-deficient phenotype of Chlamydomonas reinhardtii Ac-208 results from a frame-shift mutation in the nuclear gene encoding preapoplastocyanin. J Biol Chem 268:7832-7841
Boynton-Gillham laboratory, Duke University, 1993
Chloroplast construct containing E. coli recA with an N-end deletion, flanked by Chlamydomonas atpA 5′ and rbcL 3′; 1.97 kb in BSKS+
host strain: XL1-Blue
amp resistant
Boynton-Gillham laboratory, Duke University, 1993
Chloroplast expression cassette, 0.62 kb in BSSK+, with 5′ and 3′ ends of psbA, and NcoI/XbaI sites for insertion of any coding sequence in frame
host strain: XL1-Blue
amp resistant
Boynton-Gillham laboratory, Duke University, 1993
Chloroplast cassette, aadA flanked by psbA 5′ and 3′ regions; ca. 1.5 kb in BSKS+
host strain: XL1-Blue
amp resistant
Boynton-Gillham laboratory, Duke University, 1993
aadA flanked by psbA 5′ and atpB 3′; 3.6 kb in BSKS+
host strain: XL1-Blue
amp resistant
P-578 cpDNA EcoRI 6 (10 kb)
$30.00
$30.00
From Jean-David Rochaix, 1993
Chloroplast DNA EcoRI 6 (10 kb) in pBR328; contains psbC 3′, psaC, and rpoC1b; P-583 is the same insert in a different vector
host strain: XL1-Blue
amp resistant
From Jean-David Rochaix, 1993
Chloroplast DNA EcoRI 13 (6.0 kb) in pBR328; contains psbA exons 1-4 and extreme 3′ end of chlN
host strain: unknown
amp resistant
P-581 cpDNA EcoRI 23 or 24
$30.00
$30.00
From Jean-David Rochaix, 1993
Thought to be chloroplast DNA Eco RI fragment 23; 3.4 kb in pBR328;
could be Eco 24 instead
host strain: unknown
amp resistant
From Jean-David Rochaix, 1993
Chloroplast DNA EcoRI 28 (2.0 kb) in pCRI; internal to ORF1995
host strain: unknown
kanamycin resistant
P-583 cpDNA EcoRI 6 (10 kb)
$30.00
$30.00
From Jean-David Rochaix, 1993
Chloroplast DNA EcoRI 6 (10 kb) in pCRI; contains psbC 3′, psaC, and rpoC1b; P-578 is the same insert in pBR328
host strain: unknown
kanamycin resistant
P-584 cpDNA EcoRI 7 (8.7 kb)
$30.00
$30.00
From Jean-David Rochaix, 1993
Chloroplast DNA EcoRI 7 (8.7 kb) in pCRI; contains ccsA, psbZ, psbM, rps14, rps7, and the 5′ end of atpE
A gel run in December 1993 suggested that this insert also includes the adjacent fragment EcoRI 25.
host strain: unknown
kanamycin resistant
From Steve Surzycki, 1993
Chloroplast DNA BglII-SalI (4.3 kb) in pUC18; contains 3′ end of rps3 and 5′ end of rpoC2
host strain: JM83
amp resistant
Fong SE, Surzycki SJ (1992) Chloroplast RNA polymerase genes of Chlamydomonas reinhardtii exhibit an unusual structure and arrangement. Curr Genet 21:485-497
From Steve Surzycki, 1993
Chloroplast DNA PstI-BglII (3.1 kb) in pUC8; contains psbL, petG, and 5′ portion of rps3; subclone of chloroplast DNA fragment BglII 8
host strain: JM83
amp resistant
Fong SE, Surzycki SJ (1992) Chloroplast RNA polymerase genes of Chlamydomonas reinhardtii exhibit an unusual structure and arrangement. Curr Genet 21:485-497
P-593 Gulliver K probe
$30.00
$30.00
From Patrick Ferris, 1994
Gulliver K (left end); 4.1 kb HindIII in pUC13; used as a probe for the Gulliver transposon
host strain: unknown
amp resistant
Ferris PJ (1989) Characterization of a Chlamydomonas transposon, Gulliver, resembling those in higher plants. Genetics 122:363-377
P-594 Gulliver N probe
$30.00
$30.00
From Patrick Ferris, 1994
Gulliver N (right end); EcoRI/HindIII 0.6 kb in pUC13; contains ca. 130 bp of non-Gulliver DNA,= probe C for MT- locus
host strain: unknown
amp resistant
Ferris PJ (1989) Characterization of a Chlamydomonas transposon, Gulliver, resembling those in higher plants. Genetics 122:363-377
P-597 cpDNA PstI 22 (2.3 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1994
Chloroplast DNA PstI 22 (2.3 kb) in BSKS+, intergenic region between rpoC1a and rpoC1b
host strain: JM83
amp resistant
P-603 cpDNA PstI 12 (7.5 kb)
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1994
Chloroplast DNA PstI 12 (7.5 kb) in BSKS+; contains tscA, chlN, and psbA exons 1 and 2
host strain: JM83
amp resistant
Boynton-Gillham laboratory, Duke University, 1994
Chloroplast DNA BamHI 17 (800 bp) from CC-503 cw92 in pTZ18R; contains 3′ end and UTR of ccsA
host strain: DH5 alpha
amp resistant
P-611 OEE2 cDNA
$30.00
$30.00
From Steve Mayfield, 1994
OEE2 cDNA, 1.4 kb EcoRI fragment in pUC19
host strain: JM83
amp resistant
Mayfield SP, Rahire M, Frank G, Zuber H, Rochaix JD (1987) Expression of the nuclear gene encoding oxygen-evolving enhancer protein 2 is required for high levels of photosynthetic oxygen evolution in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 84:749-753
P-612 OEE2 genomic
$30.00
$30.00
From Steve Mayfield, 1994
OEE2 genomic, EcoRI/KpnI fragment in pUC19
host strain: JM83
amp resistant
Mayfield SP, Rahire M, Frank G, Zuber H, Rochaix JD (1987) Expression of the nuclear gene encoding oxygen-evolving enhancer protein 2 is required for high levels of photosynthetic oxygen evolution in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 84:749-753
From Pete Lefebvre, 1994
CRY1 marker; RPS14 gene, ca. 4 kb EcoRI fragment in pUC119
Derived by the following steps: pCRY1-1 was constructed by using PCR to amplify the 3.6 kb genomic region containing the entire CRY1-1 transcription unit and 250 bp of upstream sequence. The 3.6 kb product was cloned into pUC119 digested with XbaI and SacI. Plasmid pRS2-1 was constructed by amplifying a 1073 bp region of the RBCS2 promoter and cloning the PCR product into pUC119 deigested with SmaI and dT tailed with Taq polymerase. pJN1 was constructed by inserting the 3.0 kb BfrI-EcoRI fragment of pCRY1-1 into the HincII site of pRS2-1. pJN4 was constructed by ligating the 474 bp Alw26I-ApaI fragment of pCRY1-1 to the 7.2 kb ApaI-BamHI fragment of pJN1.
Lefebvre included the following suggestions on use of this plasmid:
The protocol we use for transformation is included in the Nelson et al. paper. As you will notice in the paper, the RBCS2 promoter was deleted and inverted during the PCR steps of constructing pJN4, but it still works well for transformation.
The key point to be stressed is the need to determine the appropriate killing concentration for emetine for your strain under your conditions. Emetine is variable in potency between batches, and it is light sensitive. You should determine, by replica plating, the killing concentration for your strain, and then attempt your selection on a concentration 2-3x that needed to kill all of your wild-type. We grow our cells on plates in the light, but not very strong light (about 15 inches away from a two-bulb fluorescent shop light). Make your plates only a few days in advance, and keep in the dark until use.
You might want to try selection in minimal media as well as acetate-containing media. Expression from the RBCS2 promoter is maximal in light in media without acetate.
host strain: DH4 alpha
amp resistant
Nelson JA, Savereide PB, Lefebvre PA (1994) The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol Cell Biol 14:4011-4019
P-619 cytochrome c6 5′ UTR
$30.00
$30.00
From Sabeeha Merchant, 1994
845 bp of cytochrome c6 5′ UTR, in BSKS-; sequence sufficient to drive copper-dependent expression
host strain: DH5 alpha
amp resistant
Hill KL, Li HH, Singer J, Merchant S (1991) Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J Biol Chem 266:15060-15067
P-620 cytochrome c6 5′ UTR
$30.00
$30.00
From Sabeeha Merchant, 1994
120 bp of cytochrome c6 5′ UTR in BSKS-; sequence sufficient to drive copper-dependent expression
host strain: DH5 alpha
amp resistant
Hill KL, Li HH, Singer J, Merchant S (1991) Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J Biol Chem 266:15060-15067
- «Previous Page
- 1
- …
- 3
- 4
- 5
- 6
- 7
- …
- 15
- Next Page»