Plasmids
P-628 NIC7 genome, pnic7.9
$30.00
$30.00
From Patrick Ferris, 1995
NIC7 genomic, pnic7.9
host strain: unknown
amp resistant
Ferris PJ (1995) Localization of the nic-7, ac-29 and thi-10 genes within the mating-type locus of Chlamydomonas reinhardtii. Genetics 141:543-549
P-640 NIT1 genomic
$30.00
$30.00
From Ola Sodeinde, 1995
Wild-type genomic NIT1 sequence marked with small insertion in intron 9; vector is a deletion derivative of pBR322
host strain: unknown
amp resistant
From Ola Sodeinde, 1995
Wild-type genomic NIT1 clone, minus the promoter and a portion of the first exon, marked with a small insertion in intron 9; derived from P-640
host strain: unknown
amp resistant
From Ola Sodeinde, 1995
Wild-type genomic NIT1 sequence, minus the four terminal 3′ exons and the 3′ untranslated region, and marked with a small insertion in intron 9
host strain: unknown
amp resistant
Boynton-Gillham laboratory, Duke University, 1997
E. coli aadA gene flanked by C. reinhardtii rps7 5′ and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Bam (7.4 kb) in
pUC8. In Fargo et al. (1998) this was incorrectly listed as P-665.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1997
E. coli aadA gene flanked by C. reinhardtii rps4 5′ and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Bam (7.4 kb) in
pUC8.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1997
E. coli aadA gene flanked by C. reinhardtii atpB 5′ (with no Shine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Bam (7.4 kb) in pUC8.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1997
Chloroplast DNA Bam-HindIII (4.8 kb) in pUC18, 16S rRNA through 5′ part of 23S rRNA; contains spectinomycin resistance mutation spr-u-1-6-2, from CC-110, in 3′ end of 16S rRNA; same insert as P-228 but in XL1 blue host strain
host strain: XL1-Blue
amp resistant
More information
Strain: CC-110 spr-u-1-6-2 mt+
Insert: BamHI-HindIII (4.8 kb) from BamHI 11/12
Genes present: 16S rRNA, 23Ss rRNA (5′ end)
Vector: pUC18
Bacterial host strain: XL1 blue
Selectable marker: amp-r, white on X-gal
Origin: Scott Newman in Boynton/Gillham lab, 1989; transferred to XL1 blue by Barbara Randolph-Anderson, 1995
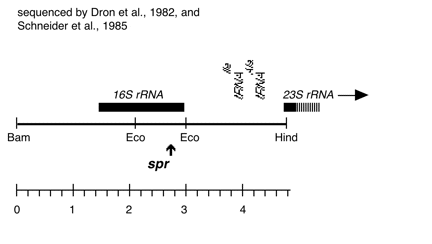
This is the same insert as P-228, but in XL1 blue
The spr-u-1-6-2 mutations is an A-> G change at bp 1123 in the C. reinhardtii 16S rRNA gene which causes loss of an AatII restriction site and confers high level spectinomycin resistance. See Harris et al., Genetics 123, 281-292 (1989). Used in rRNA transformation experiments by Newman et al., Genetics 126, 875-888 (1990).
P-668 cpDNA DCMU4 er-u-1a
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1997
psbA exon 5 to 23S rRNA 3′, with DCMU4 and er-u-1a mutations; Eco-SalI (5.7 + 1.2 kb) in BSKS
host strain: XL1-Blue
amp resistant
Boynton-Gillham laboratory, Duke University, 1997
E. coli aadA gene flanked by C. reinhardtii rps4 5′ (with no Shine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Bam (7.4 kb) in pUC8.
host strain: XL1-Blue
amp resistant
P-681 nuclear aadA cassette
$30.00
$30.00
Boynton-Gillham laboratory, Duke University, 1997
Nuclear aadA cassette; aadA (E. coli) flanked by RBCS2 5′ and 3′; SacI-Kpn (1.35 kb) in pBluescript KS II (-)
host strain: XL1-Blue
amp resistant
Cerutti H, Johnson AM, Gillham NW, Boynton JE (1997) A eubacterial gene conferring spectinomycin resistance on Chlamydomonas reinhardtii: integration into the nuclear genome and gene expression. Genetics 145:97-110
pSP108 nuclear ble cassette
$30.00
$30.00
From Saul Purton, 1997
ble (phleomycin resistance gene from S. hindustanus) flanked by RBCS2 5′ and 3′
host strain: XL1-Blue
amp resistant
Stevens DR, Rochaix JD, Purton S (1996) The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol Gen Genet 251:23-30
Boynton-Gillham laboratory, Duke University, 1998
E. coli aadA gene flanked by C. reinhardtii atpE 5′ and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli aadA gene flanked by C. reinhardtii atpE 5′ (with no Shine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli aadA gene flanked by C. reinhardtii atpB 5′ (with canonical Shine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli aadA gene flanked by C. reinhardtii rps4 5′ (with canonical Shine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli uidA (GUS) gene flanked by C. reinhardtii atpB 5′ (with canonical Shine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli uidA (GUS) gene flanked by C. reinhardtii rps4 5′ (withShine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli uidA (GUS) gene flanked by C. reinhardtii atpB 5′ and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli uidA (GUS) gene flanked by C. reinhardtii rps4 5′ and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Boynton-Gillham laboratory, Duke University, 1998
E. coli uidA (GUS) gene flanked by C. reinhardtii atpB 5′ (no Shine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli uidA (GUS) gene flanked by C. reinhardtii rps4 5′ (with putative S-D sequence eliminated) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli aadA gene flanked by f1 phage Gene VII 5′ (no S-D) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli aadA gene flanked by f1 phage Gene VII 5′ (with Shine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli uidA (GUS) gene flanked by f1 phage Gene VII 5′ (with no Shine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
Boynton-Gillham laboratory, Duke University, 1998
E. coli uidA (GUS) gene flanked by f1 phage Gene VII 5′ (with Shine-Dalgarno sequence) and rbcL 3′; inserted downstream of the atpB gene but in the opposite orientation; used to transform an atpB deletion mutant. Eco-Xho in pUC18.
host strain: XL1-Blue
amp resistant
Fargo DC, Zhang M, Gillham NW, Boynton JE (1998) Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol Gen Genet 257:271-282
From David Stern, 1998
E. coli uidA flanked by C. reinhardtii chloroplast petD 5′ and rbcL 3′; = David Stern’s pDG2
From David Stern, 1998
E. coli aadA flanked by C reinhardtii chloroplast petD 5′ and rbcL 3′; = David Stern’s pDAAD
pSP124S ble cassette
$30.00
$30.00
From Saul Purton, University College London, September 2006
ble construct in which introns were inserted into the RBCS2 5′ region to improve expression
host strain: DH5 alpha
amp resistant
Lumbreras V, Stevens D, Purton S (1998) Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J 14:441-447
pSK.KmR aphA-6
$30.00
$30.00
From Saul Purton, University College London, September 2006
aphA-6 selectable marker flanked by psbA 5′ and rbcL 3′;
host strain: DH5 alpha
amp resistant
Bateman JM, Purton S (2000) Tools for chloroplast transformation in Chlamydomonas: expression vectors and a new dominant selectable marker. Mol Gen Genet 263:404-410
- «Previous Page
- 1
- …
- 4
- 5
- 6
- 7
- 8
- …
- 15
- Next Page»